Research Mission
The goal of OMAL’s research is to understand molecular mechanisms driving carcinogenesis and the reversal of this process through treatment, by utilization and advancement of optical microscopy techniques. These techniques include multiplex immunofluorescence, increasing spatial resolution, image quantification, and in vivo and in silico modeling of tumor processes. In collaboration with Dr. David Wink (CCR, NCI), we are applying these capabilities to understand how novel treatment regimens can reverse tumorigenesis driven by inflammation and immune-suppression.
Research Descriptions
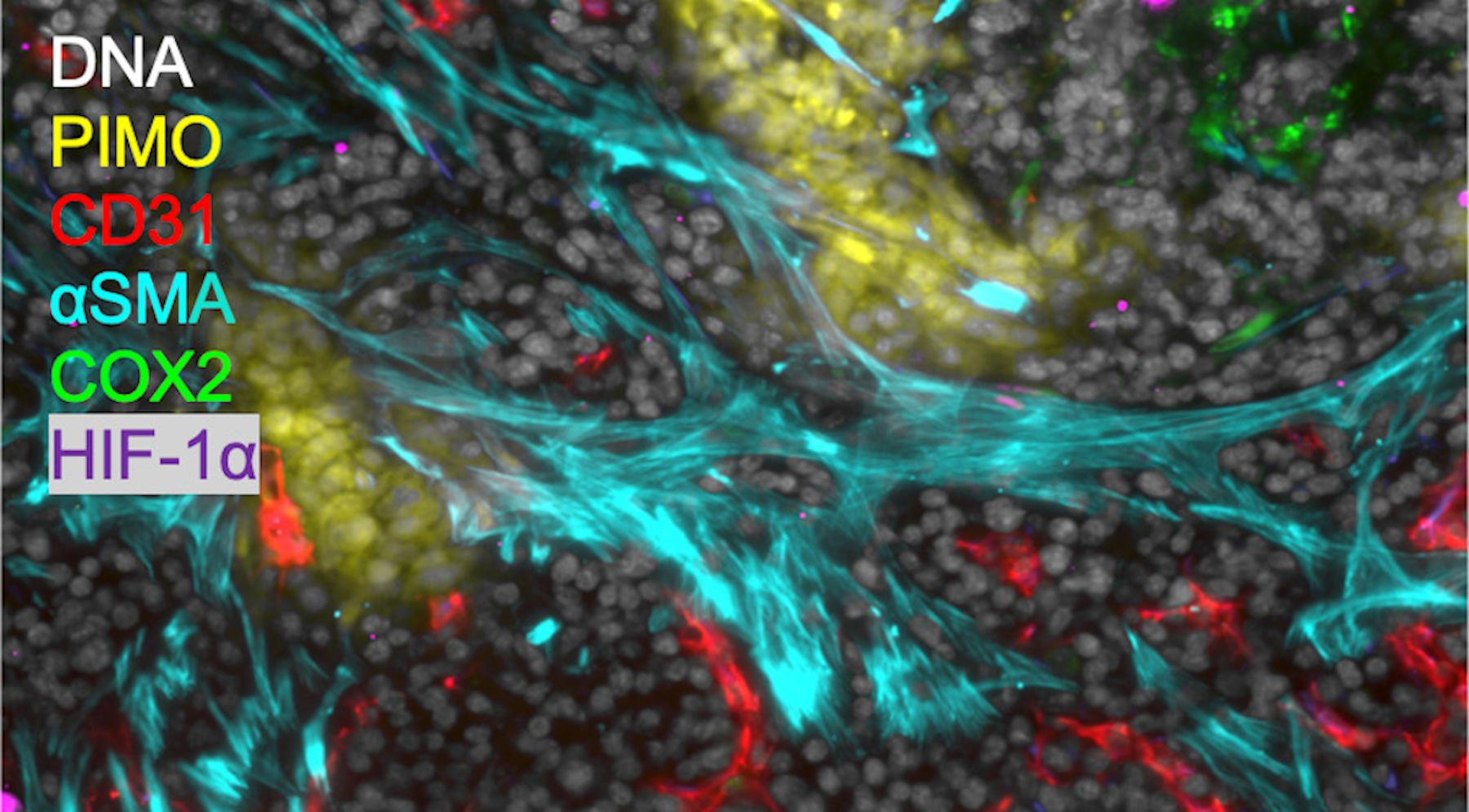 MIF used to study hypoxia and inflammation in mouse tumor tissue. Scale bar: 50 μm.
MIF used to study hypoxia and inflammation in mouse tumor tissue. Scale bar: 50 μm.Multiplexed Immunofluorescence (MIF)
We are utilizing multiplex immunofluorescence (MIF) imaging technologies employing more than 30 phenotypic markers to spatially map out 2D and 3D cellular organization in thin and thick tissue sections to elucidate the understanding of the interplay of immune cells in the immuno-suppressive tumor microenvironment (TME) caused by inflammation. We do this through two methods (1) CO-Detection by indEXing (CODEX) (Akoya Biosciences, LLC) provides a standard panel of over 20 antibodies to do MIF imaging and (2) an in-house MIF protocol developed to use antibodies pertinent to our biological questions.
Thus, our in-house MIF protocol looks at markers targeting epithelial to mesenchymal transition (metastatic phenotype), proliferation, hypoxia, cancer stem-like cell markers (treatment resistant phenotype), immune cells (immunosuppressive phenotype), stromal cells and angiogenesis. By chemically bleaching fluorescence dyes, up to 5 cycles of immunofluorescence labeling can be undertaken with 3 different antibodies in each cycle, enabling 15 antibodies to be imaged in the same sample through our in-house MIF protocol.
We have also shown that 2D MIF applied to thin tissue sections often misses the additional complexity of 3D cellular interactions and quantitative accuracy is poor at the individual cell level because cells are not intact. Thus, we have extended our technology for 3D investigation of thick tissue sections to 50 microns depth, enabling the 3D spatial relationships of cells to be determined. With our 2D and 3D multiplex immunofluorescent technique, we can investigate how different immune cell types interact within a cancerous region of the TME, as well as discover the nature of cancer stem-like cell niches.
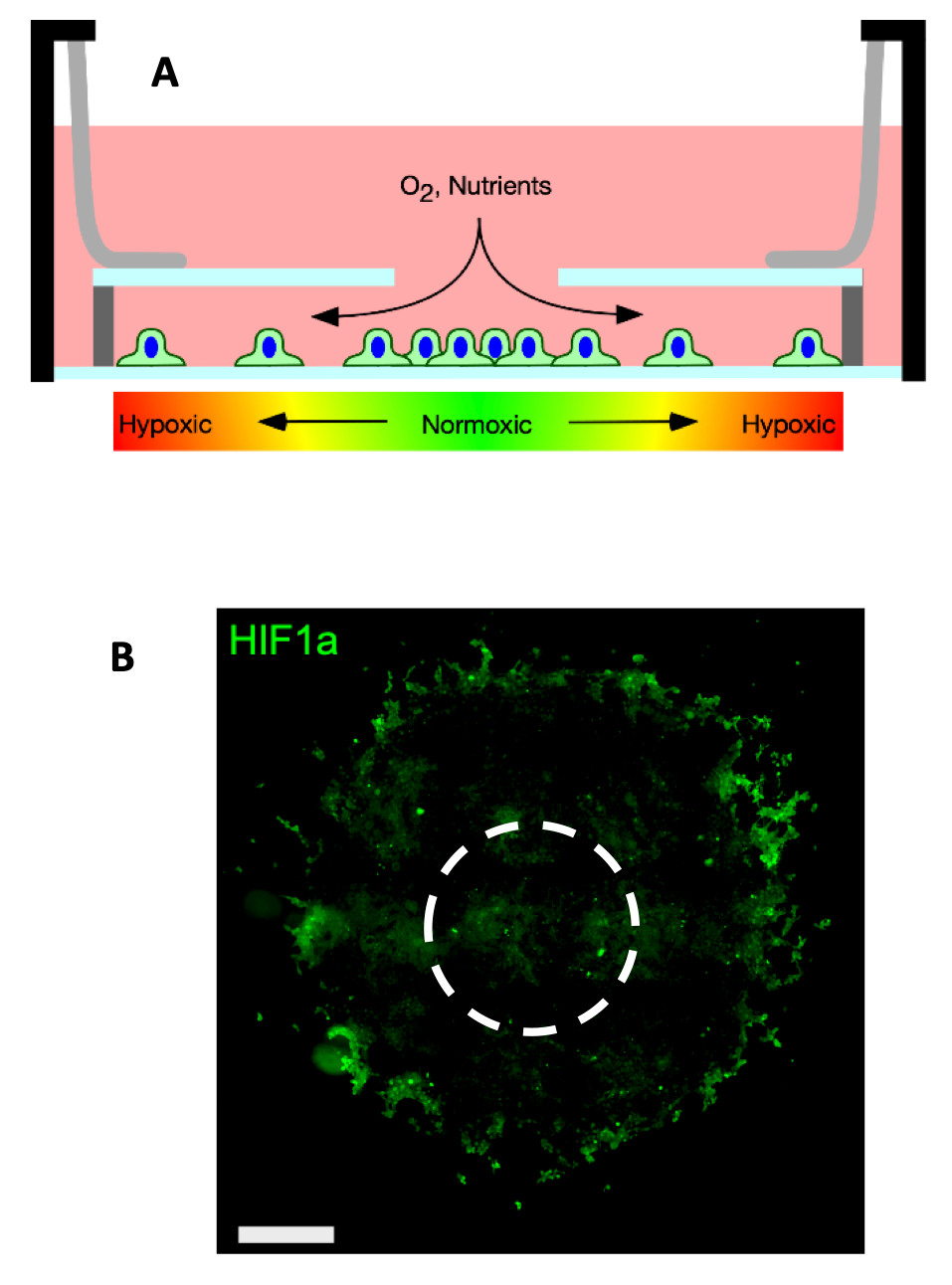 A: Schematic of the hypoxia-gradient chamber. Cells cultured in the chamber generate hypoxic gradients via consumption of oxygen diffusing through the opening the in the chamber top. B: 4T1 cells cultured in the chamber for several days form disks. The image shows the disk immunofluorescence labeled against hypoxia inducible factor 1-alpha (HIF1-⍺ – a marker of intercellular hypoxia). Cells are present inside the HIF1⍺ positive ring but are unlabeled. The dashed circle represents the hole in the chamber top. Scale bars.
A: Schematic of the hypoxia-gradient chamber. Cells cultured in the chamber generate hypoxic gradients via consumption of oxygen diffusing through the opening the in the chamber top. B: 4T1 cells cultured in the chamber for several days form disks. The image shows the disk immunofluorescence labeled against hypoxia inducible factor 1-alpha (HIF1-⍺ – a marker of intercellular hypoxia). Cells are present inside the HIF1⍺ positive ring but are unlabeled. The dashed circle represents the hole in the chamber top. Scale bars. Tumorigenesis & Inflammation From The Inside Out
We aim to understand tumorigenesis and inflammation by investigating the tumor microenvironment in a spatially resolved manner. The tumor microenvironment (TME) is complex and heterogenous, consisting of gradients of oxygen and diffusing soluble molecules and the co-existence of multiple cell types, which can affect a number of cellular processes and signaling pathways. We developed a 2D in vitro model of the TME that recapitulates the organization of cancer cells in a rapidly growing tumor in which hypoxic regions arise when the normal vasculature is no longer sufficient to oxygenate the entire tumor mass. We have used REECs, live cell microscopy, and multiplexed immunofluorescence methods to investigate the proinflammatory signaling of inducible nitric oxide synthase (NOS2) and cyclooxygenase (COX2) in the 4T1 syngeneic mouse model of triple-negative human breast cancer. High expression of both proteins strongly predicts poor survival in ER- breast cancer. We quantified the spatial distribution of biomarkers such as HIF-1α, EMT markers, and Nos2/Cox2 of 4T1 cells in the REECs and other 4T1 tumor systems.
Moreover, Nos2 cells were localized in hypoxic regions, as anticipated since hypoxia and nutrient deprivation drive Nos2 expression. Furthermore, we demonstrated that stimulated ANA1 macrophages extend the normoxic region in these gradients through increased Nos2 expression and nitric oxide production, and that 4T1 cells and macrophages segregate into distinct regions, with 4T1 cells migrating to the source of oxygen and nutrients. Ongoing studies are 1) investigating how different immune cell types interact in the tumor microenvironments before and after treatment with anti-inflammatory drugs and 2) discovering the cellular organization of cancer stem-like cell niches.
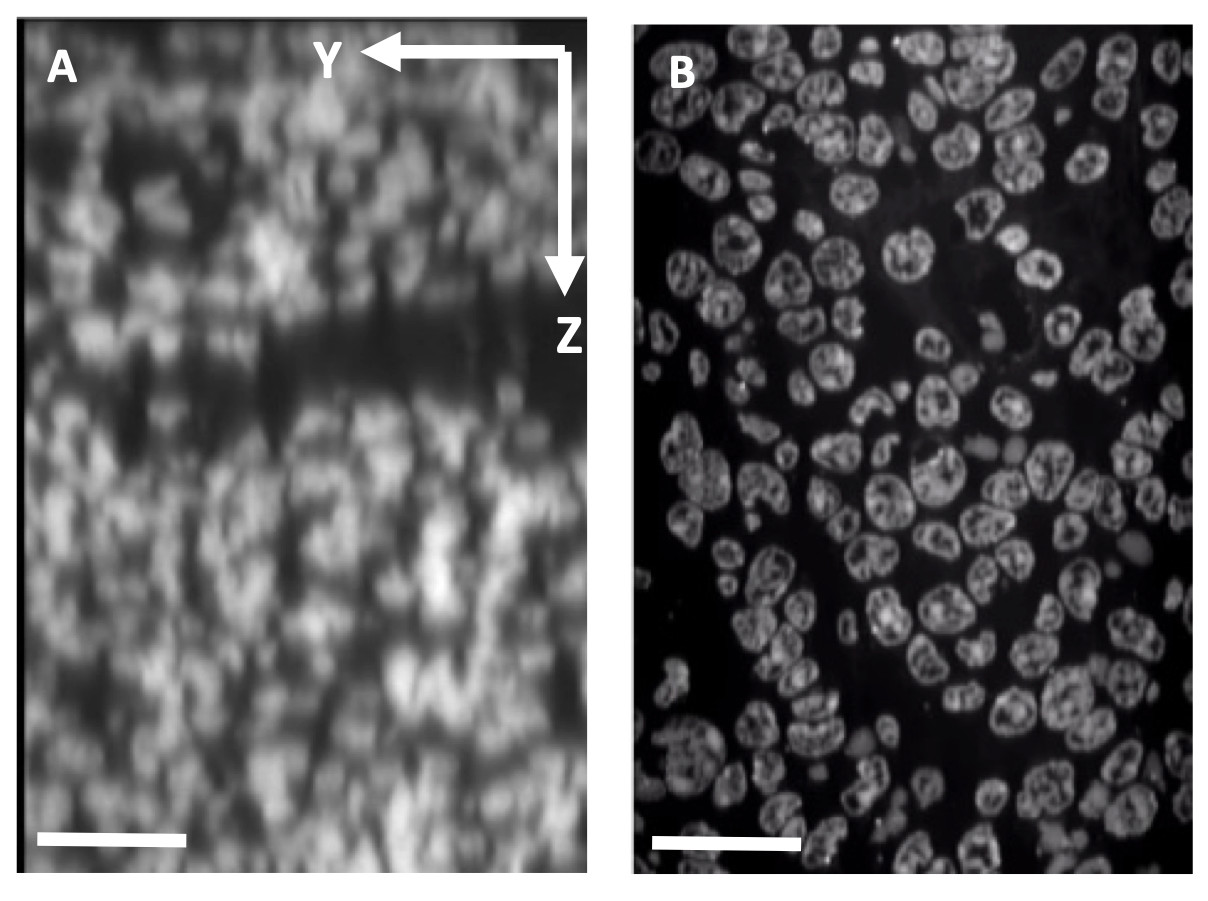 Tissue expansion to increase the effective spatial resolution. A: Confocal microscope depth slice through 4T1 mouse tumor tissue showing Yopro-1 labeled nuclei. Scale bar: 25 μm. B: Equivalent slice after expansion. Scale bar: 100 μm.
Tissue expansion to increase the effective spatial resolution. A: Confocal microscope depth slice through 4T1 mouse tumor tissue showing Yopro-1 labeled nuclei. Scale bar: 25 μm. B: Equivalent slice after expansion. Scale bar: 100 μm.Tissue Clearing & Expansion Microscopy
We follow recently established techniques (e.g., expansion microscopy, tissue clearing, etc.) to study diffraction-limited biological questions and improve both antibody and optical penetration within our samples. Recent major developments in tissue clearing—a variety of sample preparation techniques designed to reduce light scattering in samples—have profoundly increased resolution and tissue penetration in optical microscopy. Examples of these novel techniques include organic solvent clearing methods (e.g., vDISCO) which enable whole animal fluorescence imaging, as well as simple immersion clearing methods (e.g., Ce3D) which enhance transparency of medium-sized samples (e.g., small embryos, tissue sections <200 µm); we have customized Ce3D for several applications.
Alternative approaches to mitigate the effects of light scatter utilize tissue-hydrogel hybrids and share a common workflow in which a robust hydrogel scaffold is polymerized throughout a biological sample while anchoring structures of interest to the polymer network, prior to the removal of unwanted material (e.g., lipids) by enzymatic digestion or detergent treatment. Multiple hydrogel-embedding techniques, generally referred to as expansion microscopy (ExM), have also enabled super-resolution imaging with conventional microscopes. With ExM, samples can be expanded in three dimensions (~4-fold linearly) due to the swellable scaffold and reduced mechanical strength of biological material. We partner with NCI investigators to apply hydrogel-tissue chemistry to their specific biological systems; this in turn allows us to better grasp the intricacies and dynamics of these sample preparation methods and generalize and/or optimize these techniques across model organisms and imaging applications.
